Thread by Noah F. Greenwald (@NoahGreenwald@genomic.social)
- Tweet
- Nov 18, 2021
- #Biology
Thread
I’m super excited to share our work, out today @NatureBiotech: a new deep learning algorithm to accurately identify cells in imaging data! Our method works across image platforms, tissue types, and species with no fine-tuning required 🧵 www.nature.com/articles/s41587-021-01094-0
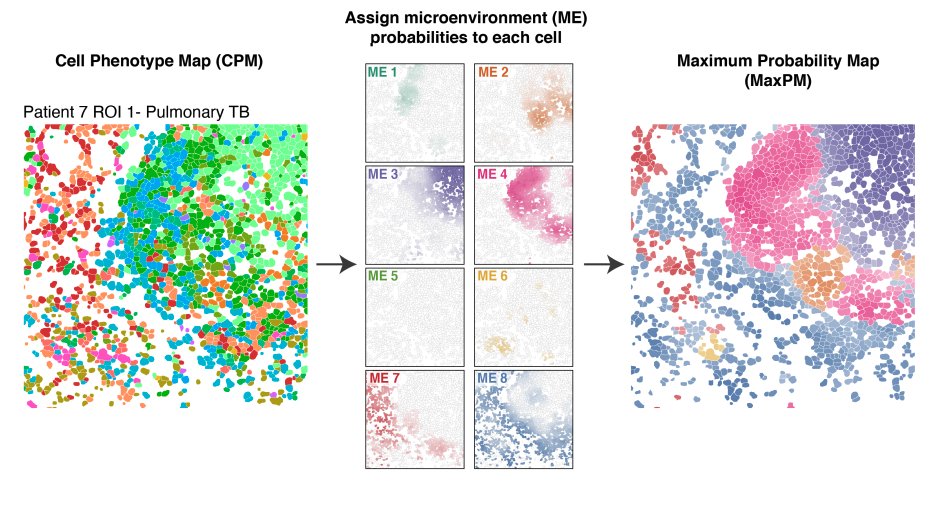
Imaging data lets us understand the spatial organization of cells in health and disease, such as in this awesome work on Tuberculosis infection from @ErinMcCaffrey14. However, these cool analyses can’t be performed on raw images; you first need to uniquely identify each cell 2/x
This process of identifying each cell in an image is known as cell segmentation, and it turns out to be a pretty hard problem, especially in tissues. One reason it’s challenging is that we don’t have large, high-quality datasets to train tissue segmentation algorithms 3/x
Creating segmentation training data is extremely time intensive. We developed a new tool, DeepCell Label, for annotating image data. We then used crowdsourcing to enable hundreds of annotators to manually label every cell in thousands of images 4/x
github.com/vanvalenlab/deepcell-label
github.com/vanvalenlab/deepcell-label
The result of this process is TissueNet, a segmentation training dataset with more than 1 million manually annotated cells. TissueNet is substantially larger than all previous datasets combined, with data from 6 different imaging platforms and 9 different tissue types 5/x
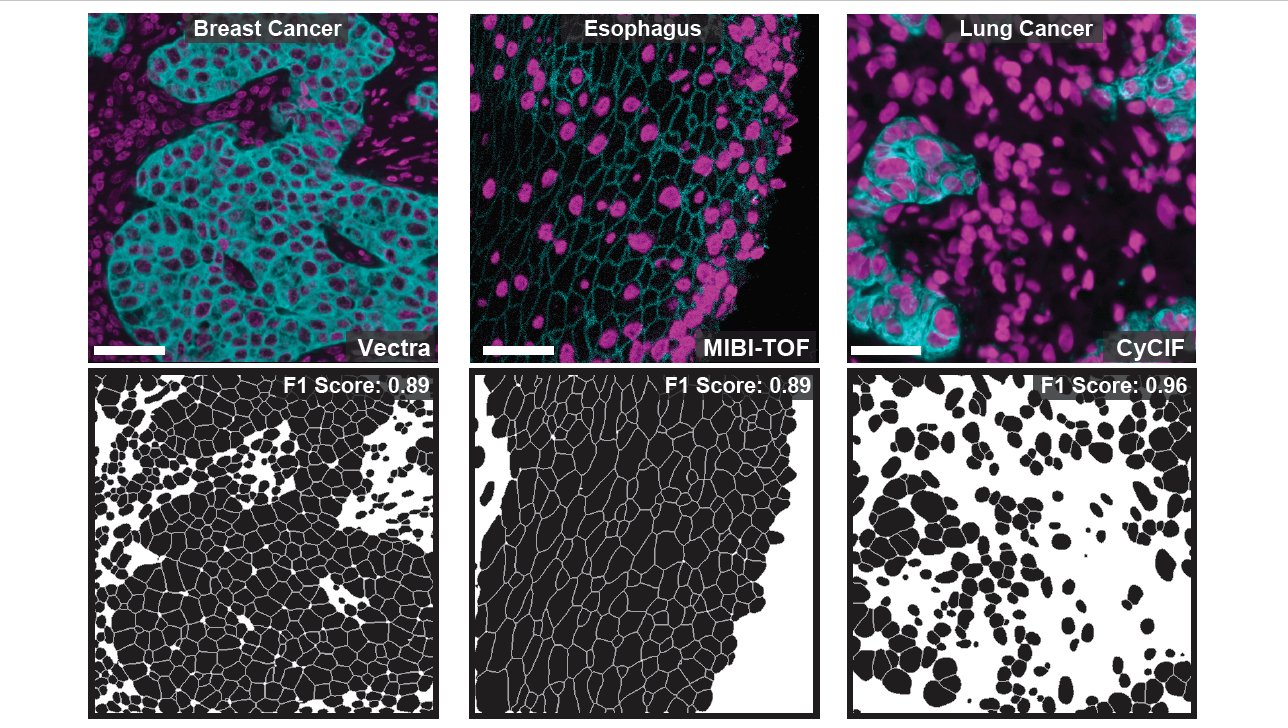
We used TissueNet to train our deep learning algorithm, Mesmer. Mesmer can accurately segment cells across the full diversity of tissue types and microscope platforms present in TissueNet without any manual intervention or parameter adjustment required 6/x
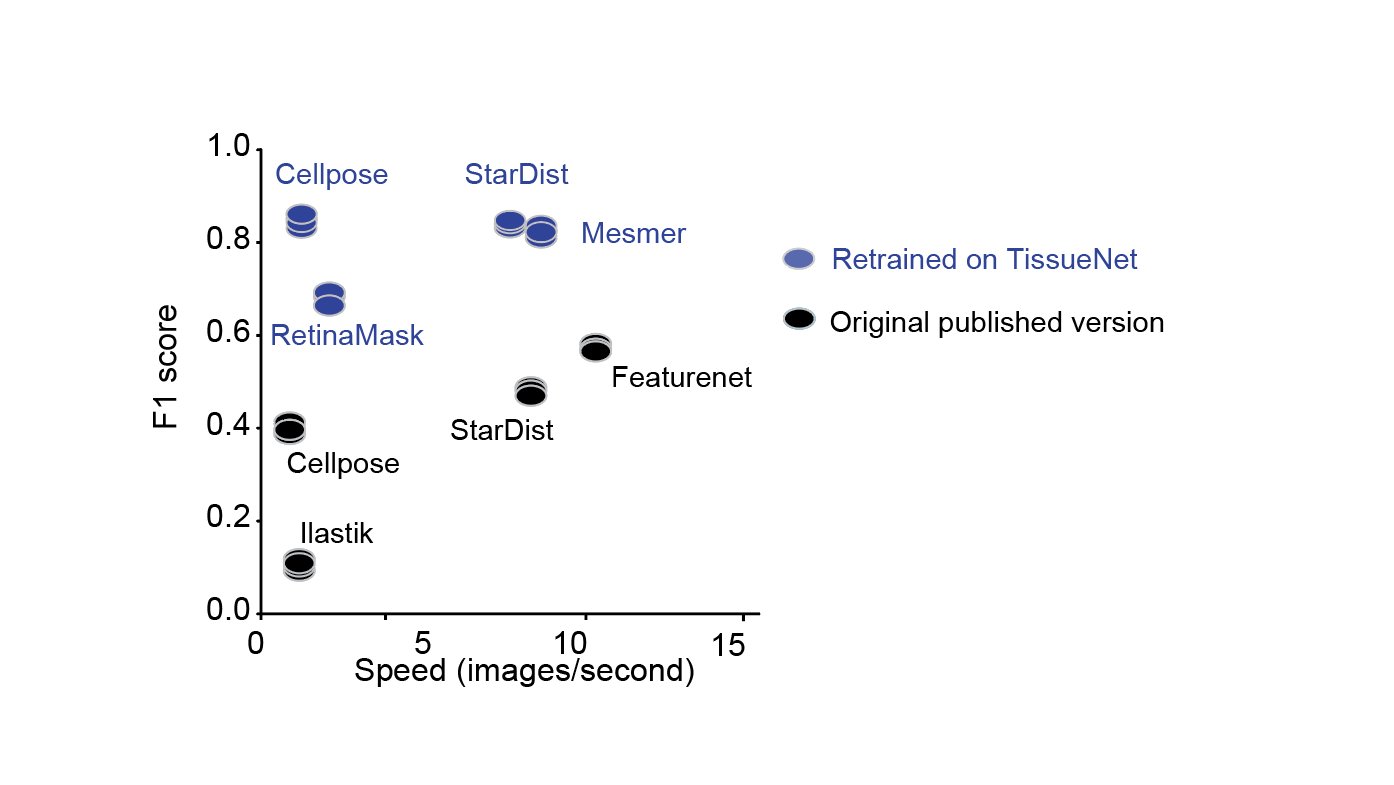
Furthermore, when we retrained previously published algorithms using TissueNet, we found that CellPose and StarDist achieved excellent performance, demonstrating the wide utility the TissueNet dataset will have as a resource for model training and development 7/x
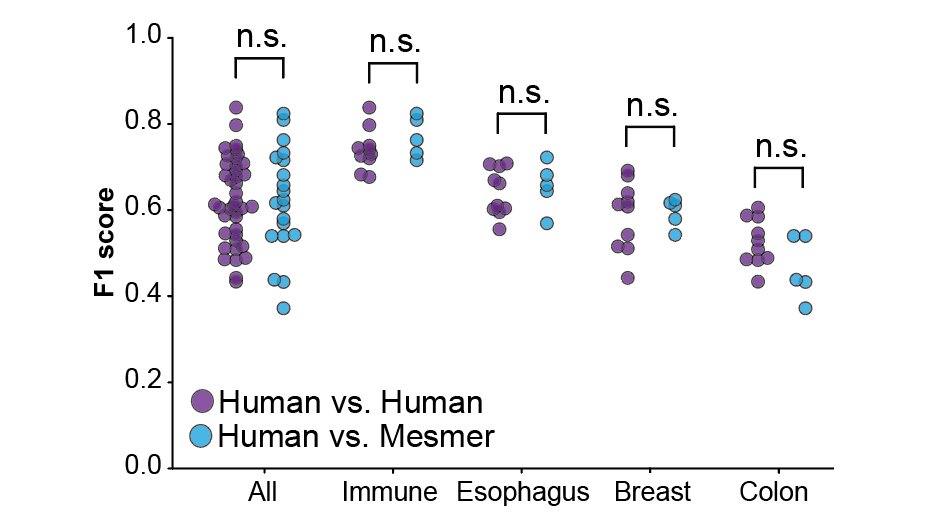
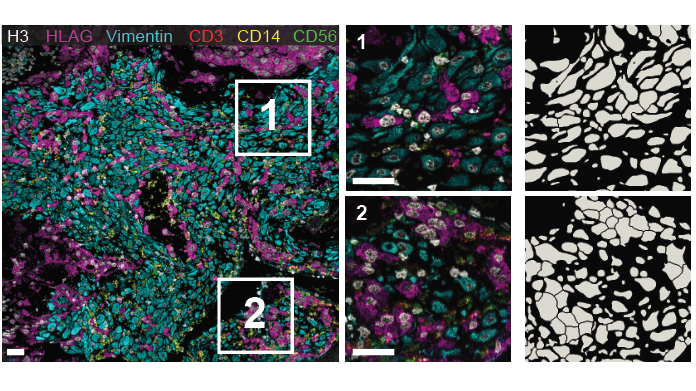
Next, we compared the segmentations from human annotators with the predictions from Mesmer. We found no significant differences between the human annotators and Mesmer, indicating we had achieved human-level performance, a first for tissue segmentation! 8/x
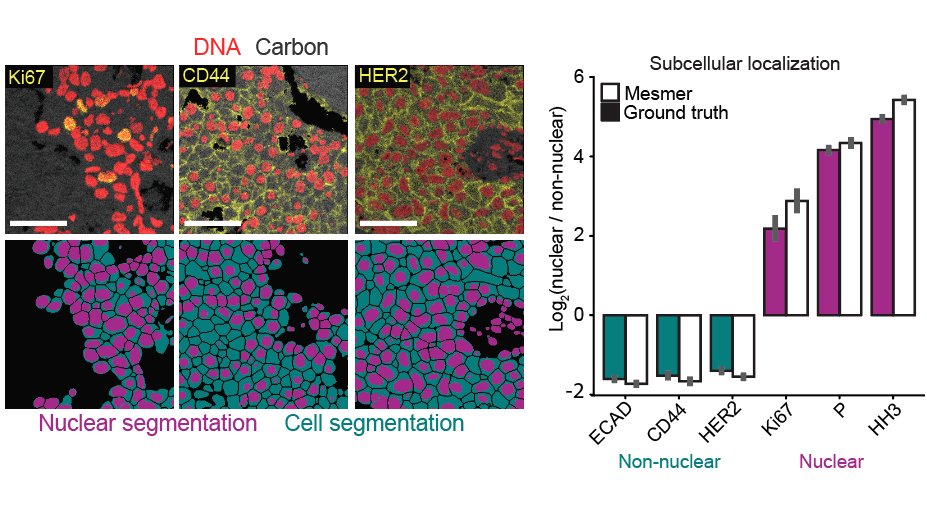
Mesmer identifies both the nucleus and cytoplasm for each cell, unlike previous nucleus-only models. We highlighted some of the cool stuff you can do with this new data, such as accurately predicting the subcellular localization of imaging signal within tissue sections 9/x
Finally, we showed how Mesmer can be fine-tuned on specific datasets. Techniques like MIBI-TOF can generate images with dozens markers, and we retrained Mesmer to take advantage of this rich information to guide segmentation decisions at the maternal-fetal interface 10/x
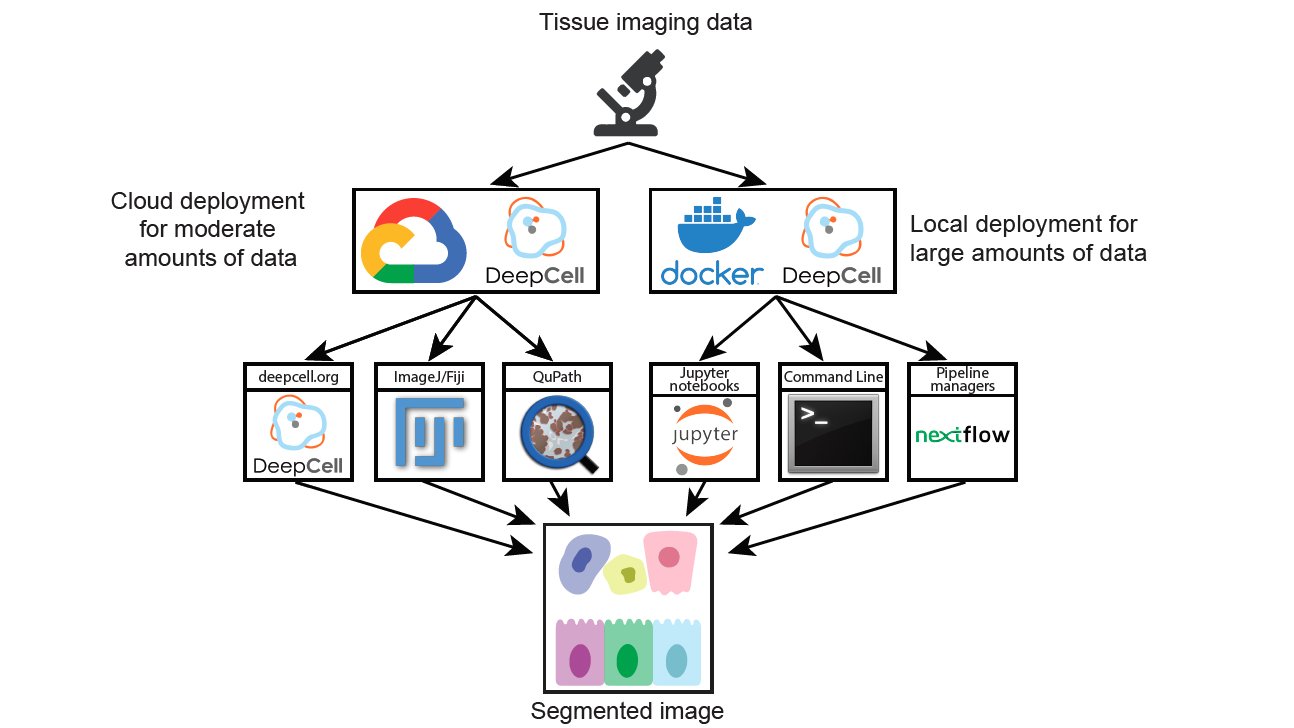
Fancy algorithms that no one can use don’t provide much value to the community. To ensure that Mesmer is as useful as possible, we’ve harnessed DeepCell to create a variety of different ways for you to use Mesmer to analyze your data 11/x
To get started, you can upload your images to Deepcell.org, which hosts the Mesmer model for anyone to use. You can also look at our intro repo, which has a full description of the command line tools, notebooks, and plugins to run Mesmer 12/x github.com/vanvalenlab/intro-to-deepcell
All of the gory details, plus lots of juicy stuff that couldn’t fit in this thread, can be found in the paper. Give it a read, and try Mesmer out for yourself! 13/x
Finally, I wanted to thank my irreplaceable co-author @BiologistGeneva; the whole team that made this possible including @DallasMoenNews, @alex_l_kong, @adam_kagel, and @realwillgraf; along with my stellar mentors @leeat_keren, @MikeAngeloLab, and @davidvanvalen 14/end